Seurat v4 vs Seurat v5
Here I summarize the most relevant conceptual and structural changes introduced in Seurat v5 compared to Seurat v4, with an emphasis on how these changes impact single-cell RNA-seq analysis workflows.
This notebook is intended to frame the new conceptual model behind Seurat v5 and its alignment with current best practices.
Versioning note
This overview reflects Seurat v5.x behavior as of early 2026. Minor interface details may evolve across releases, but the core design principles described here are stable.
Unless explicitly stated otherwise, examples and concepts discussed here refer primarily to RNA assays. Multimodal (RNA + ATAC) considerations are noted where relevant.
What does Seurat v5 introduce?
Seurat v5 introduces an internal reorganization of the data object and the analysis workflow with the following goals:
- Improve scalability for large datasets
- Facilitate multimodal analyses
- Allow methodological flexibility in normalization and integration
- Reduce unnecessary data duplication within the object
These changes reflect a shift toward more modular, explicit, and comparable workflows.
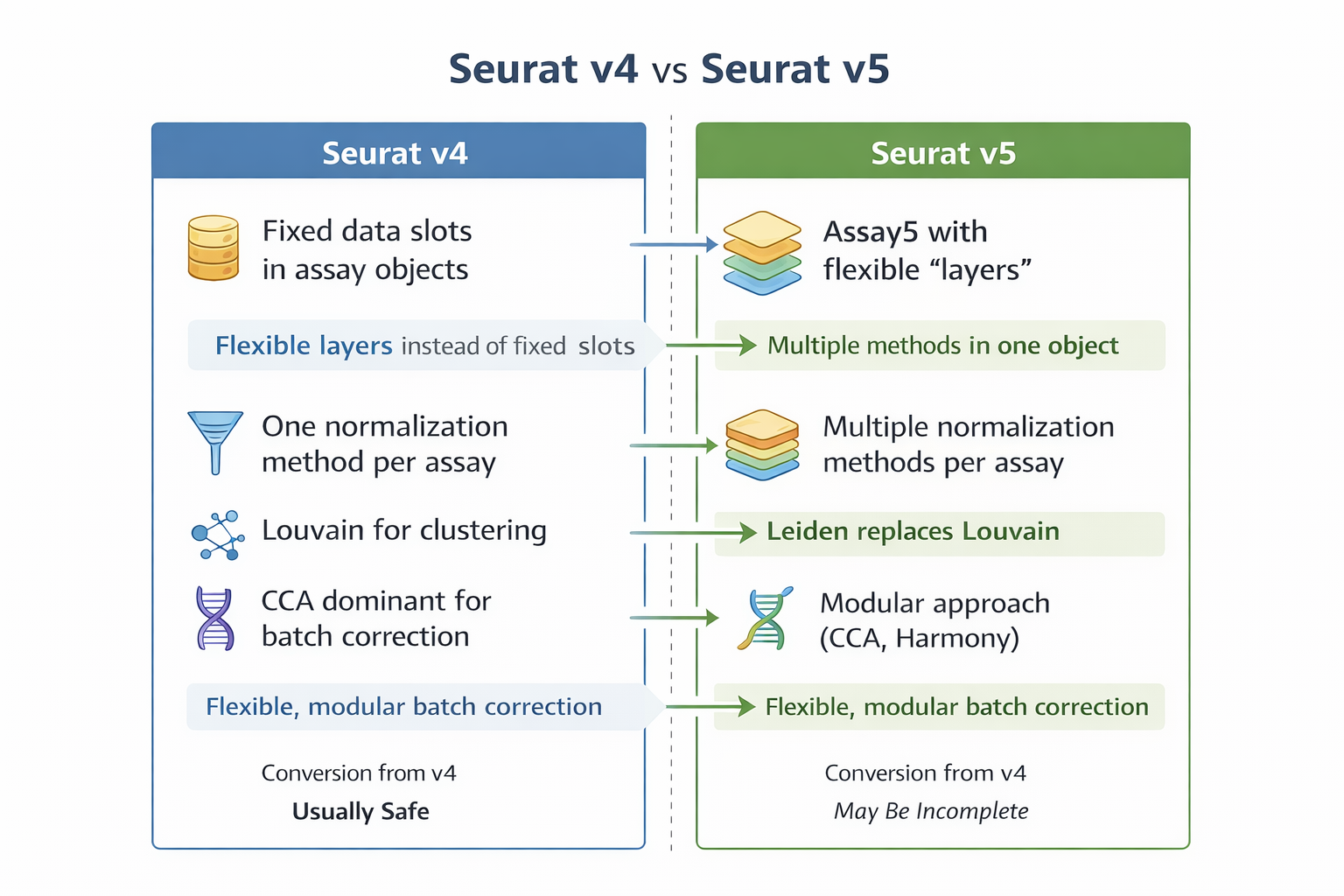
Main structural change: from ´Assay´ to ´Assay5´
Seurat v4
- Each
Assaycontains fixed slots:countsdatascale.data
- A single normalization strategy per assay
Seurat v5
- Introduces
Assay5and the concept of layers - Allows multiple representations of the same data within a single assay
- Explicit separation between:
- raw data
- normalized data
- transformed data
This design avoids object duplication and enables alternative analytical strategies without recomputing the entire workflow.
The concept of layers in Seurat v5
A layer represents a specific data representation within an assay. Common examples include:
countslognormsctransformpearson_residuals
This structure provide us a key of advantages, now we can:
- Compare different normalization methods within the same object
- Modify downstream analyses without recomputing upstream steps
- Enable methodological benchmarking
Normalization: conceptual differences between v4 and v5
Seurat v4
- Common workflows use
NormalizeData()(methods: LogNormalize, CLR, RC) orSCTransform()as a separate variance-stabilizing normalization framework - Normalization choice is typically exclusive and assay-bound
Seurat v5
- The layer-based data model allows multiple normalized representations to coexist within the same assay
- External normalization strategies (e.g. scran, shifted-log) can be stored as layers
- Pearson residuals are supported as first-class outputs in residual-based workflows
- Downstream analyses explicitly reference the layer being used, rather than inheriting assumptions from the assay
Additional workflow-relevant changes
-
Feature selection
Expanded from predominantly variance-based HVGs toward analytical, model-based approaches such as Pearson residuals and Deviance, while retaining variance-based methods. -
Dimensionality reduction
Decoupled from the assay and explicitly defined by layer, improving clarity and flexibility. -
Clustering
Shift from Louvain algorithm to Leiden as the default method, althought Louvain is still supported. This change push to get a better partition stability and faster convergence.Related resources reviewed on 2025, but still on practice:
Ver presentación: Metodos de agrupamiento en datos multiome
- Differential analysis
Growing emphasis on pseudobulk and donor-aware modeling, aligned with recent benchmarking studies, while retaining cell-level testing options
Summary comparison
| Aspect | Seurat v4 | Seurat v5 |
|---|---|---|
| Data object | Fixed-slot Assay | Assay5 with layers |
| Data representations | One per assay | Multiple layers |
| Normalization | LogNormalize or SCTransform | Multiple stored representations (e.g. scran, residuals) |
| Feature selection | Variance-based HVGs | Pearson residuals and Deviance |
| Dimensionality reduction | Assay-coupled | Layer-defined |
| Default clustering | Louvain | Leiden |
| DGE | Mostly cell-level | Pseudobulk and mixed models |
What changes if I migrate today?
| Aspect | If you stay on Seurat v4 | If you migrate to Seurat v5 |
|---|---|---|
| Object structure | Fixed slots per assay | Assay5 with multiple layers |
| Normalization | One method per assay | Multiple normalization strategies within the same assay |
| Method comparison | Requires recomputation | Native support via layers |
| Clustering default | Louvain | Leiden |
Addendum: Signac workflows (RNA + ATAC) in Seurat v5
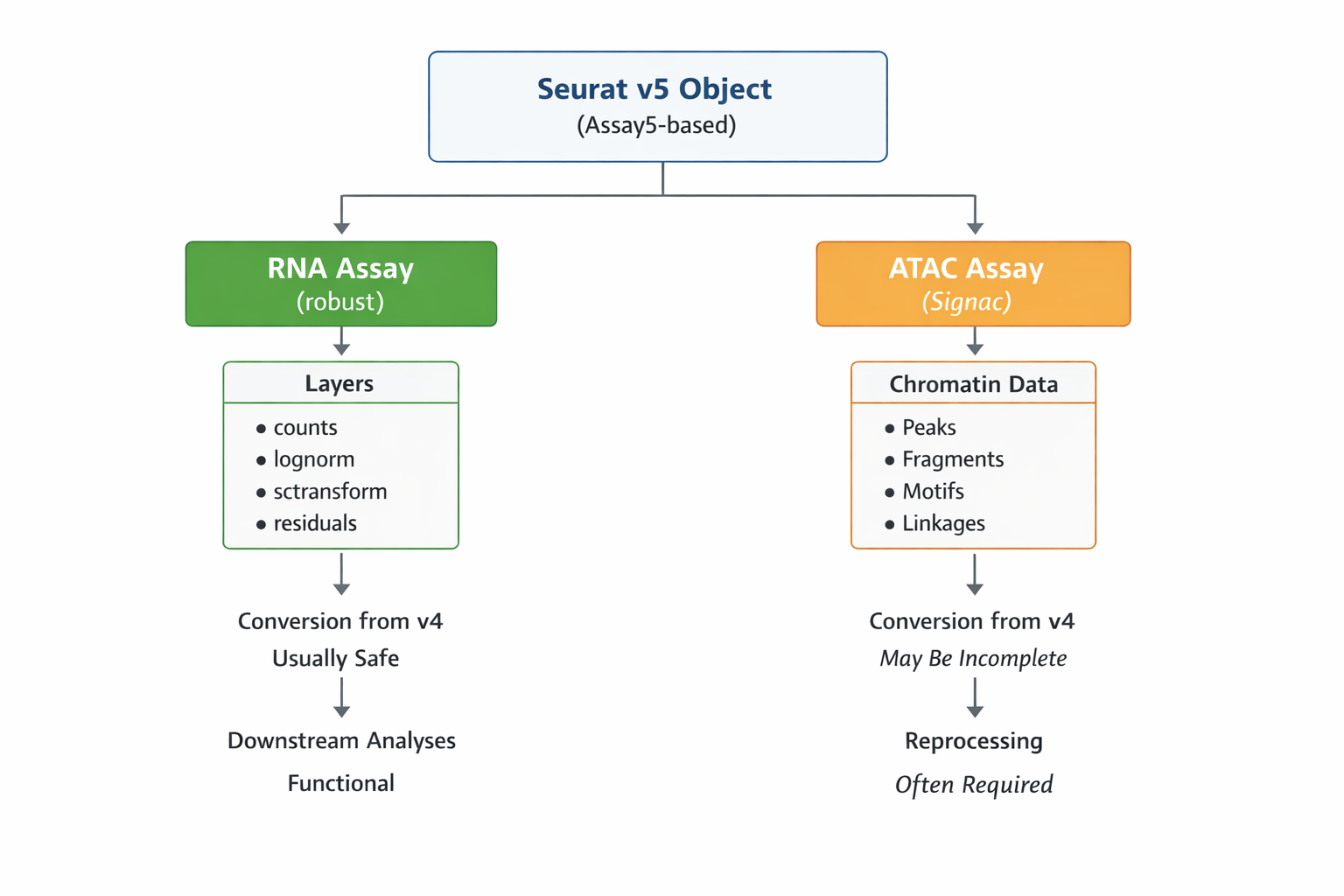
While Seurat v5 provides a unified object model for multimodal data, Signac-based chromatin workflows introduce additional constraints that are important to keep in mind during migration.
Key considerations:
-
Chromatin assays are more sensitive to object conversion than RNA assays
In particular, legacy objects created under Seurat v4 + Signac v1.x may not always retain full chromatin functionality when updated to Seurat v5. -
Assay5 does not automatically guarantee chromatin compatibility
A converted chromatin assay may exist structurally but fail downstream operations (e.g. peak-based reductions, motif analysis, or linkage steps). - Recommended practice
For multimodal projects (RNA + ATAC), especially those involving:- peak calling
- TF motif analysis
- peak–gene linkage
it is often safer to reconstruct the chromatin assay directly under Seurat v5 / Signac v2, rather than relying on object conversion.
- RNA assays are generally robust to conversion, while ATAC assays benefit from explicit reprocessing.
This distinction is particularly relevant for large multiome datasets and for workflows that rely heavily on chromatin-specific reductions and annotations.
Alignment with Seurat v5.1+ developments
The conceptual framework described in this notebook remains stable across Seurat v5 releases. However, recent versions (v5.1+) further reinforce the design principles introduced in v5.0.
These refinements include:
- More consistent handling of layers across functions
- Improved interoperability with external methods
- Clearer separation between data representation and analysis logic
I have found that Seurat v5.1+ have make a clear improvement toward transparent, modular, and benchmark-aligned workflows, rather than introducing breaking conceptual changes.
Users migrating from Seurat v4 are therefore encouraged to adopt the layer-centric mindset early, as it represents the stable direction of the framework going forward.
This notebook serves as a conceptual reference to understand these changes and to support informed decisions when designing new analyses or migrating existing projects.
Resources
-
Crowell et al. (2020) – Benchmarking scRNA-seq analysis pipelines
-
Squair et al. (2021) – Best practices for differential expression in scRNA-seq
CSC. February 2026